What Is PrimerPipeline?
PrimerPipeline: A user-friendly Windows based program enveloping an automated pipeline for microsatellite mining and primer design for de novo low coverage genomes.
With the decreasing price and widening availability of Next Generation Sequencing it is now incredibly quick and simple to produce a library of sequences for species requiring microsatellite primers. However, taking this data to fruition: primers ready to test and optimise, can be difficult for those unfamiliar with bioinformatics. Most of the pipelines on offer are in the form of a Linux based command line, for ecologists not familiar with bioinformatics, scripts or the Linux language, this can be time consuming and potentially lead to mistakes. Here we introduce PrimerPipeline, an open source, user-friendly Windows based (GUI) program that does not require any bioinformatics knowledge and allows you to find microsatellites and design primers using the best, most established methods (MISA and Primer3) but develop, display and check the results in a novel, clear and simple way.
Why Use It?
PrimerPipeline incorporates sequence trimming, microsatellite mining and microsatellite primer design. PrimerPipeline automatically makes input files, and continues onto the next step, making it much more time efficient for the user, because instead of having to run several scripts separately and make new input files for each step, you can now (once you have chosen your settings for the three steps) set the pipeline running with the touch of a button!
Useful Features
- The main feature is that the program is so simple and straightforward to use.
- Keeps you informed of its progress - PrimerPipeline has a status bar which gives you an up-to-date percentage complete for each step of the pipeline. It also counts the number of sequences read and the number of microsatellites found.
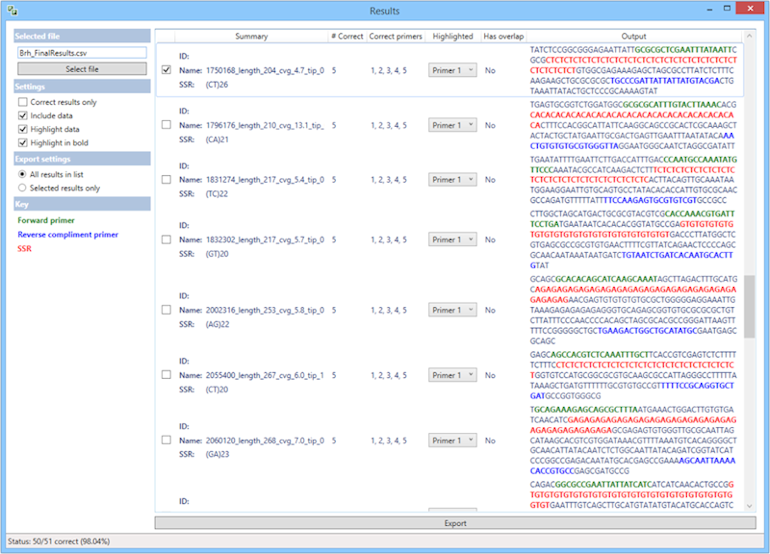
- Checks the results ensuring that Primer3 has designed the primers correctly, so that they amplify the desired microsatellite, and that there is no overlap. At the bottom it displays a percentage correct and you can tick Correct results only in the Results window to only view and export correct primers.
- From the Results window you can select individual sequences (along with their information) and export them to a new CSV file to help you select your desired primers.
- You can copy data from the Results window along the formatting (the highlighted primer and SSR) and paste into Word, PowerPoint etc., to present your data.